publications
2024
-
 SC-MIL: Supervised Contrastive Multiple Instance Learning for Imbalanced Classification in PathologyDinkar Juyal, Siddhant Shingi, Syed Ashar Javed, Harshith Padigela, Chintan Shah, Anand Sampat, Archit Khosla, John Abel, and Amaro Taylor-WeinerIn Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision (WACV), Jan 2024
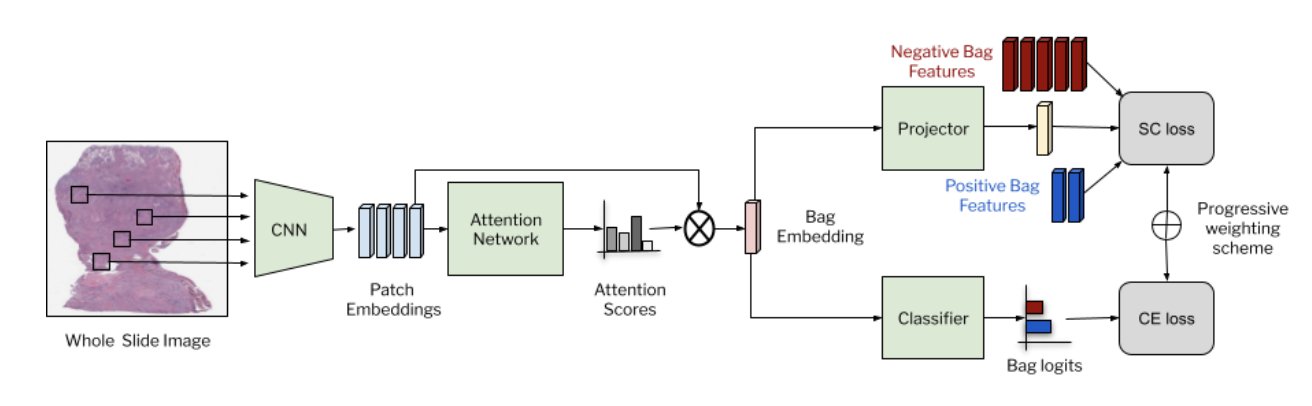
SC-MIL: Supervised Contrastive Multiple Instance Learning for Imbalanced Classification in PathologyDinkar Juyal, Siddhant Shingi, Syed Ashar Javed, Harshith Padigela, Chintan Shah, Anand Sampat, Archit Khosla, John Abel, and Amaro Taylor-WeinerIn Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision (WACV), Jan 2024Multiple Instance learning (MIL) models have been extensively used in pathology to predict biomarkers and risk-stratify patients from gigapixel-sized images. Machine learning problems in medical imaging often deal with rare diseases, making it important for these models to work in a label-imbalanced setting. Furthermore, these imbalances can occur in out-of-distribution (OOD) datasets when the models are deployed in the real-world. We leverage the idea that decoupling feature and classifier learning can lead to improved decision boundaries for label imbalanced datasets. To this end, we investigate the integration of supervised contrastive learning with multiple instance learning (SC-MIL). Specifically, we propose a joint-training MIL framework in the presence of label imbalance that progressively transitions from learning bag-level representations to optimal classifier learning. We perform experiments with different imbalance settings for two well-studied problems in cancer pathology: subtyping of non-small cell lung cancer and subtyping of renal cell carcinoma. SC-MIL provides large and consistent improvements over other techniques on both in-distribution (ID) and OOD held-out sets across multiple imbalanced settings.
- Foundation AI models predict molecular measurements of tumor purityYlaine Gerardin, Daniel Shenker, Jennifer Hipp, Natalia Harguindeguy, Dinkar Juyal, Chintan Shah, Syed Ashar Javed, Marc Thibault, Michael Nercessian, Darpan Sanghavi, and othersCancer Research, Jan 2024
- P008 Machine learning derived histological features of epithelial injury and repair in Ulcerative Colitis biopsies correlate with disease severityM Griffin, Y Zhang, C Shah, J Shamshoian, V Mountain, K Sucipto, C Jayson, and F NajdawiJournal of Crohn’s and Colitis, Jan 2024
-
 PLUTO: Pathology-Universal TransformerDinkar Juyal*, Harshith Padigela*, Chintan Shah*, Daniel Shenker, Natalia Harguindeguy, Yi Liu, Blake Martin, Yibo Zhang, Michael Nercessian, Miles Markey, Isaac Finberg, Kelsey Luu, Daniel Borders, Syed Ashar Javed, Emma Krause, Raymond Biju, Aashish Sood, Allen Ma, Jackson Nyman, John Shamshoian, Guillaume Chhor, Darpan Sanghavi, Marc Thibault, Limin Yu, Fedaa Najdawi, Jennifer A. Hipp, Darren Fahy, Benjamin Glass, Eric Walk, John Abel, Harsha Pokkalla, Andrew H. Beck, and Sean GrullonMay 2024
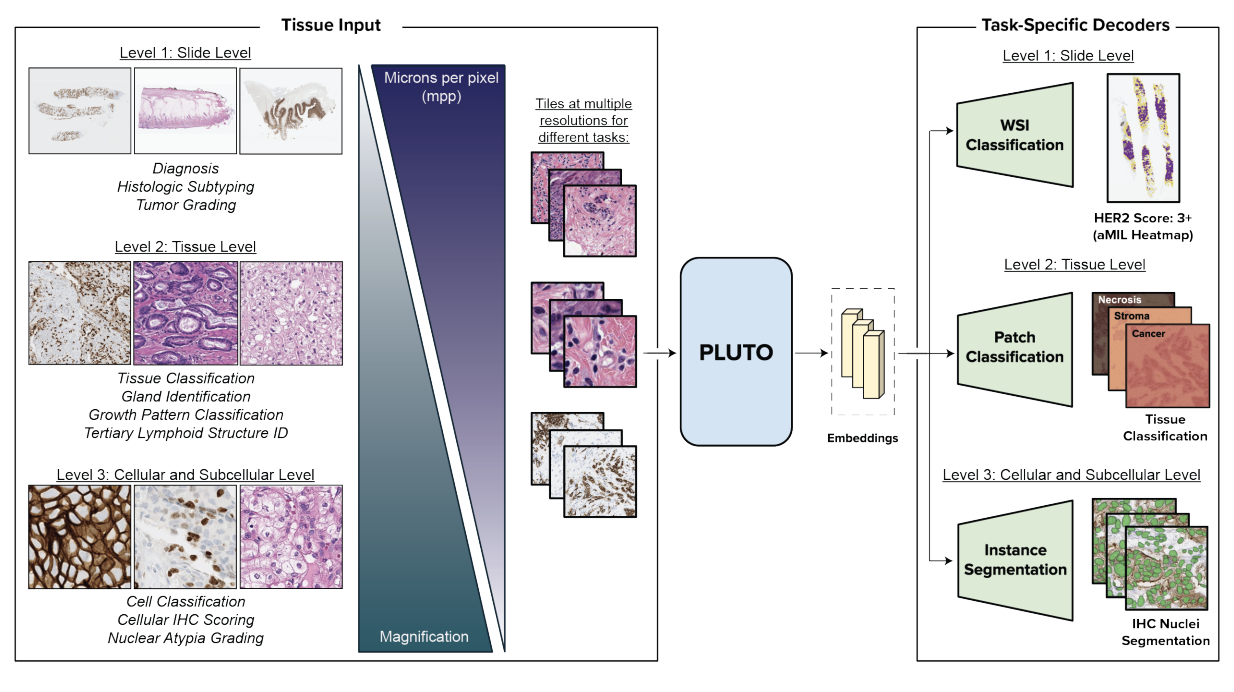
PLUTO: Pathology-Universal TransformerDinkar Juyal*, Harshith Padigela*, Chintan Shah*, Daniel Shenker, Natalia Harguindeguy, Yi Liu, Blake Martin, Yibo Zhang, Michael Nercessian, Miles Markey, Isaac Finberg, Kelsey Luu, Daniel Borders, Syed Ashar Javed, Emma Krause, Raymond Biju, Aashish Sood, Allen Ma, Jackson Nyman, John Shamshoian, Guillaume Chhor, Darpan Sanghavi, Marc Thibault, Limin Yu, Fedaa Najdawi, Jennifer A. Hipp, Darren Fahy, Benjamin Glass, Eric Walk, John Abel, Harsha Pokkalla, Andrew H. Beck, and Sean GrullonMay 2024 - Interpretability analysis on a pathology foundation model reveals biologically relevant embeddings across modalitiesNhat Le, Ciyue Shen, Chintan Shah, Blake Martin, Daniel Shenker, Harshith Padigela, Jennifer Hipp, Sean Grullon, John Abel, Harsha Vardhan Pokkalla, and Dinkar JuyalMay 2024
2023
- Synthetic DOmain-Targeted Augmentation (S-DOTA) Improves Model Generalization in Digital PathologySai Chowdary Gullapally, Yibo Zhang, Nitin Kumar Mittal, Deeksha Kartik, Sandhya Srinivasan, Kevin Rose, Daniel Shenker, Dinkar Juyal, Harshith Padigela, Raymond Biju, Victor Minden, Chirag Maheshwari, Marc Thibault, Zvi Goldstein, Luke Novak, Nidhi Chandra, Justin Lee, Aaditya Prakash, Chintan Shah, John Abel, Darren Fahy, Amaro Taylor-Weiner, and Anand SampatMay 2023
- Fully automated histological classification of cell types and tissue regions of celiac disease is feasible and correlates with the Marsh scoreMichael Griffin, Aaron Gruver, Chintan Shah, Qasim Wani, Darren Fahy, Archit Khosla, Christian Kirkup, Daniel Borders, Jackie A Brosnancashman, Angie D Fulford, and othersmedRxiv, May 2023
- ContriMix: Unsupervised disentanglement of content and attribute for domain generalization in microscopy image analysisTan H Nguyen, Dinkar Juyal, Jin Li, Aaditya Prakash, Shima Nofallah, Chintan Shah, Sai Chowdary Gullapally, Michael Griffin, Anand Sampat, John Abel, and othersarXiv preprint arXiv:2306.04527, May 2023
2022
- 552 Characteristics of the tumor microenvironment in IDH1-mutated cholangiocarcinoma patients from ClarIDHy trialH Duygu Saatcioglu, Juan Valle, Teresa Macarulla, Milind Javle, Do-Youn Oh, Lipika Goyal, Jake Conway, Janani Iyer, Fedaa Najdawi, Chintan Shah, Camelia Gliser, Susan Pandya, Scott Daigle, Ghassan Abou-Alfa, and Robin KelleyJournal for ImmunoTherapy of Cancer, May 2022
Background Somatic isocitrate dehydrogenase 1 mutations (IDH1m) convert α-ketoglutarate to the oncogenic metabolite R-2-hydroxyglutarate (2-HG). IDH1m are detected in approximately 13% of intrahepatic cholangiocarcinomas (CCAs).1 Ivosidenib, an oral inhibitor of the IDH1m protein inhibits 2-HG and restores immune response in CCA.2 We analyzed pre-treatment samples, using machine learning models to quantify histologic features of the CCA tumor microenvironment, enabling identification of correlates of IDH1m status, early disease progression (patients experienced progression or death within 1.54 months), and plasma 2-HG levels (median, 630 ng/ml).Methods A set of H&E images, including from ClarIDHy3, a phase 3 placebo controlled clinical trial of ivosidenib in IDH1m CCA, were split into training/validation (n=200) and test sets for model development. Whole slide images were annotated by GI pathologists to identify and quantify more than 500 different human interpretable features (HIFs), including cell (cancer cell, lymphocyte, macrophage, plasma cell, fibroblast) and tissue (cancer epithelium, stroma, necrosis) features. Utilizing IDH1m and wild type (WT) screening samples, multivariate logistic regression models were trained to predict IDH1m status. P-values were calculated by univariate logistic regression and corrected for multiple comparisons via adjustment for FDR.Results A HIF-based multivariate model discriminated between IDH1m and WT CCA (AUC, 0.83; 95% CI, 0.74-0.92). IDH1m was associated with a lower proportion of lymphocytes throughout the tumor (OR, 0.64; P<0.01; FDR P=0.022), and higher proportion of fibroblasts (OR, 1.8; P<0.01; FDR P=0.023) and lower proportion of plasma cells in the stroma (OR, 0.68; P<0.01; FDR P=0.032 ) (figure 1A). In a subset of samples, CD3 and CD8 staining showed reduced T-lymphocyte infiltration patterns in IDH1m (n=5) samples relative to IDH1 WT (n=19) (figure 1B). Early disease progression of enrolled ClarIDHy patients (ivosidenib n=61, placebo n=38) was associated with a higher proportion of macrophages (OR, 1.70; P<0.01; FDR P=0.08) and a lower proportion of tumor infiltrating lymphocytes (OR, 0.63; P<0.01; FDR P=0.08), (figure 2A). When correcting for treatment effect, the proportion of lymphocytes in the tumor were still associated with improved PFS (P=0.011). Consistent with previously published data2, high 2-HG levels were associated with lower numbers of tumor infiltrating lymphocytes (OR, 0.63; P=0.011; FDR P=0.08) (figure 2B).Conclusions Quantitative histologic evaluation suggests that pre-treatment IDH1m CCA samples have a colder tumor microenvironment relative to IDH1 WT CCA, with an immunosuppressive tumor microenvironment being associated with early progression. Results from this analysis support exploration of combination with immune checkpoint inhibitors.Trial Registration NCT02989857ReferencesBoscoe AN, Rolland C, Kelley RK. Frequency and prognostic significance of isocitrate dehydrogenase 1 mutations in cholangiocarcinoma: a systematic literature review. J Gastrointest Oncol. 2019;10(4):751–765. doi: 10.21037/jgo.2019.03.10.Wu MJ, Shi L, Dubrot J, et al. Mutant IDH Inhibits IFN?-TET2 signaling to promote immunoevasion and tumor maintenance in cholangiocarcinoma. Cancer Discov. 2022;12(3):812–835. doi: 10.1158/2159-8290.CD-21-1077.Abou-Alfa GK, Macarulla T, Javle MM, et al. Ivosidenib in IDH1-mutant, chemotherapy-refractory cholangiocarcinoma (ClarIDHy): a multicentre, randomised, double-blind, placebo-controlled, phase 3 study. Lancet Oncol. 2020;21(6):796–807. doi: 10.1016/S1470-2045(20)30157-1. Epub 2020 May 13. Erratum in: Lancet Oncol. 2020 Oct;21(10):e462.Ethics Approval This study was done according to the International Conference on Harmonisation of Good Clinical Practice guidelines and the principles of the Declaration of Helsinki. Approval from the institutional review board and international ethics committee was obtained at each study site. Patients provided written, informed consent before participating in the study.Abstract 552 Figure 1 Tumor microenvironment of IDH1m vs IDH1 WTTumor microenvironment of IDH1m CCA compared to IDH1 WT at screening. (A) 163 screening samples, including IDH1m (n=138) and IDH1 WT (n=25) subjects were analyzed by machine learning of histological features. Samples deemed by a panel of GI pathologist to be extrahepatic as a best response were excluded from the analysis. IDH1m status in CCA was associated with lower proportions of lymphocytes in the tumor (Upper Row), higher proportions of fibroblasts in the stroma (Middle Row), and lower proportions of plasma cells in the stroma (Bottom Row). Tumor includes cancer epithelium and stroma tissues in the whole sections. Uncorrected P values are displayed on the Figures (B) Further analysis of a subset of screening samples (n=5 IDH1m, n=19 IDH1 WT) by CD3 and CD8 staining was performed. Representative whole slide biopsy H&E images indicating lower proportions of lymphocytes in the IDH1m CCA tumor via machine learning-derived predictions (Upper Row; Lymphocytes are indicated with dark green marker overlay. MLO=Machine Learning Overlay, representative image for CD3 immunohistochemistry (Middle Row), and representative image for CD8 immunohistochemistry (Bottom Row).Abstract 552 Figure 2 Differences in CCA tumor microenvironment.Differences in CCA tumor microenvironment based on early disease progression and pre-treatment plasma 2-HG levels. (A) Pre-treatment screening samples from 99 (ivosidenib cohort n=61, placebo cohort n=38) patients treated on the ClarIDHy study were analyzed for association with early disease progression, defined as experiencing progression or death within 1.54 months (47 days) (PFS<1.54 months) Early disease progression was associated with lower proportions lymphocytes over immune cells in cancer epithelium (Upper Row) and higher proportions of macrophages (Bottom Row) over immune cells in cancer epithelium (B) Plasma 2-HG levels were available for 100 IDH1m patients, with sample groups separated based on the median plasma 2-HG level (630 ng/ml). Higher plasma 2-HG levels were associated with lower proportions of lymphocytes in CCA tumor. Uncorrected P values are displayed on the Figures (A and B)
- Self-training of Machine Learning Models for Liver Histopathology: Generalization under Clinical ShiftsJin Li, Deepta Rajan, Chintan Shah, Dinkar Juyal, Shreya Chakraborty, Chandan Akiti, Filip Kos, Janani Iyer, Anand Sampat, and Ali BehroozMay 2022
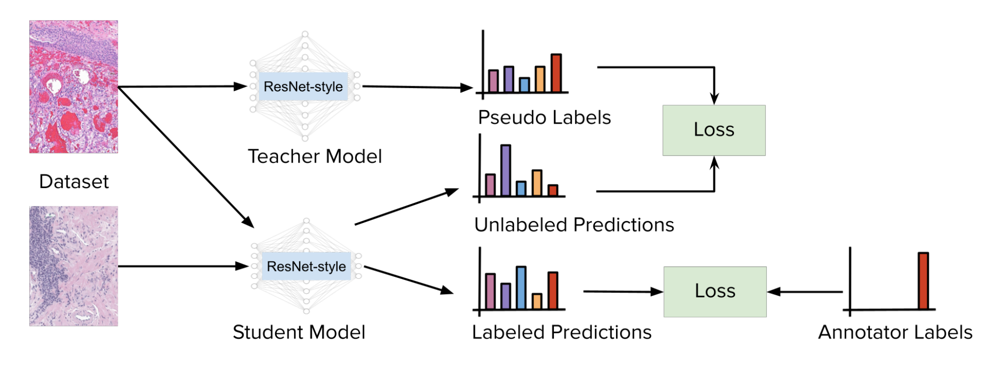
Histopathology images are gigapixel-sized and include features and information at different resolutions. Collecting annotations in histopathology requires highly specialized pathologists, making it expensive and time-consuming. Self-training can alleviate annotation constraints by learning from both labeled and unlabeled data, reducing the amount of annotations required from pathologists. We study the design of teacher-student self-training systems for Non-alcoholic Steatohepatitis (NASH) using clinical histopathology datasets with limited annotations. We evaluate the models on in-distribution and out-of-distribution test data under clinical data shifts. We demonstrate that through self-training, the best student model statistically outperforms the teacher with a 3% absolute difference on the macro F1 score. The best student model also approaches the performance of a fully supervised model trained with twice as many annotations.
2020
-
 Finding Patient Zero: Learning Contagion Source with Graph Neural NetworksChintan Shah, Nima Dehmamy, Nicola Perra, Matteo Chinazzi, Albert-László Barabási, Alessandro Vespignani, and Rose YuMay 2020
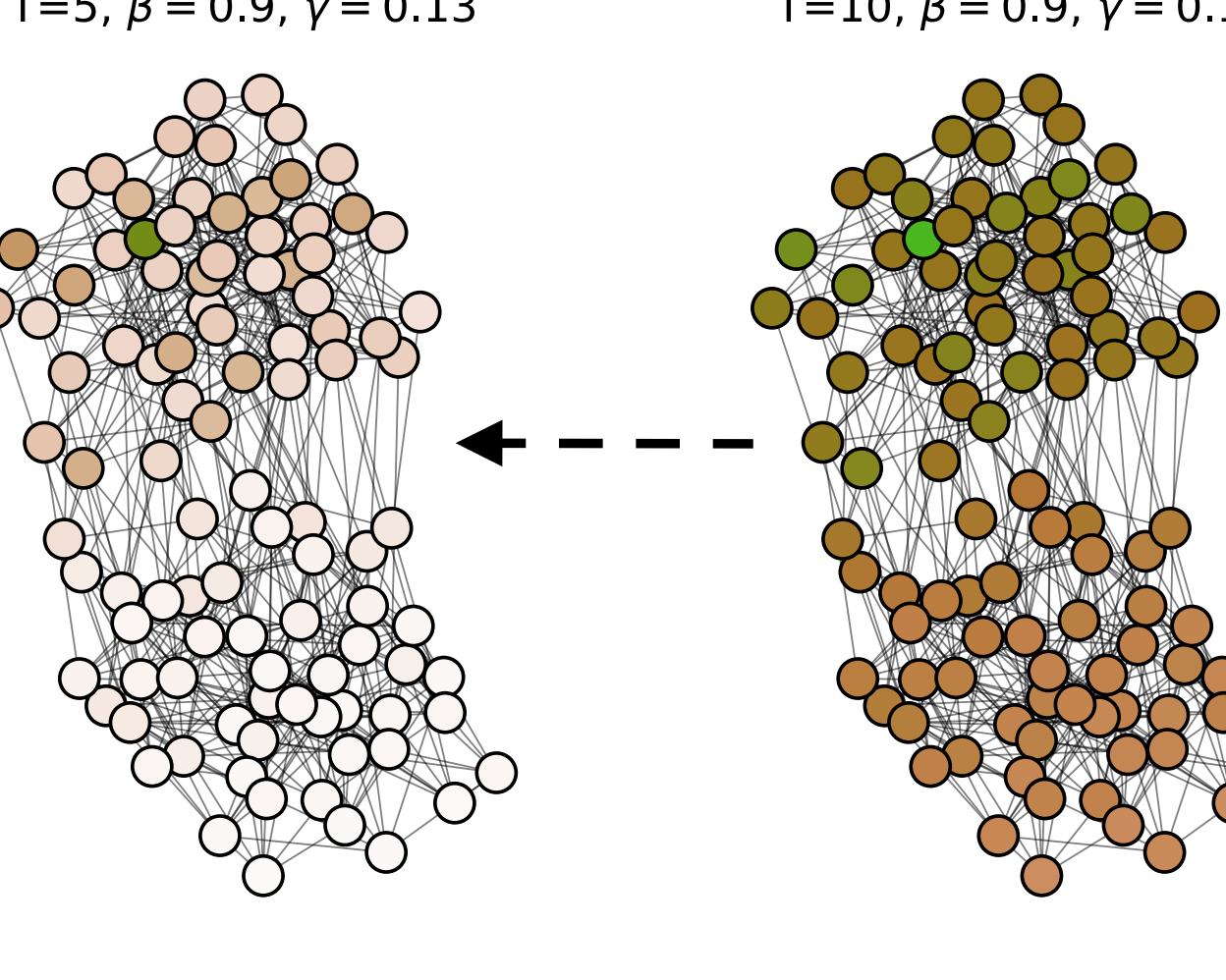
Finding Patient Zero: Learning Contagion Source with Graph Neural NetworksChintan Shah, Nima Dehmamy, Nicola Perra, Matteo Chinazzi, Albert-László Barabási, Alessandro Vespignani, and Rose YuMay 2020Locating the source of an epidemic, or patient zero (P0), can provide critical insights into the infection’s transmission course and allow efficient resource allocation. Existing methods use graph-theoretic centrality measures and expensive message-passing algorithms, requiring knowledge of the underlying dynamics and its parameters. In this paper, we revisit this problem using graph neural networks (GNNs) to learn P0. We establish a theoretical limit for the identification of P0 in a class of epidemic models. We evaluate our method against different epidemic models on both synthetic and a real-world contact network considering a disease with history and characteristics of COVID-19. % We observe that GNNs can identify P0 close to the theoretical bound on accuracy, without explicit input of dynamics or its parameters. In addition, GNN is over 100 times faster than classic methods for inference on arbitrary graph topologies. Our theoretical bound also shows that the epidemic is like a ticking clock, emphasizing the importance of early contact-tracing. We find a maximum time after which accurate recovery of the source becomes impossible, regardless of the algorithm used.